Abstract
The TP53 gene can be affected by hypomorphic/loss of function lesions occurring in diverse configurations. In addition to truncating hits, missense mutations in various hotspots may exert a dominant negative effect. Li-Fraumeni syndrome and acquired disease examples demonstrate that the presence of a wild type (WT) allele may be protective, and biallelic inactivation has the strongest leukemogenic drive. Consequently, cases with apparent biallelic TP53 lesions are associated with poor overall survival (OS) in myeloid neoplasia (MN). However, clinically applied NGS techniques currently do not always clearly identify biallelic subclones within seemingly single hit mutants, as the latter may contain subclones of various sizes. Thus, poorer than expected prognosis may be present in some cases depending on the presence and size of cryptic biallelic clones (hemizygous, homozygous, biallelic two hits) vs truly monoallelic or biclonal TP53 mosaic cases. Assessing the impact of variables surrounding TP53 mutations (TP53MT) is complex and not always feasible. Thus, the development of clinically viable algorithms may improve prognostic precision in clinical settings.
We collected clinical and molecular data from 7400 MN patients and applied a novel model to properly resolveTP53MTallelic configuration of and assess prognosis. Overall, 1285 TP53MT were found in 1010 patients. Missense mutations were the most common (74%), followed by truncating (15%), whereas 11% of the patients had concomitant missense/truncating configuration. Most missense mutations (69%) were in canonical hotspot locations. TP53MTcarriers presented with a variety of phenotypes and displayed worse OS compared to TP53WT (HR=2.7; 95%CI 2.5-3). Following the traditional criteria for estimating allelic hits1,36% of cases were classified as single hits, while 64% exhibited double hit configuration. Overall, carriers of TP53 double hits (either missense and/or truncated) had a worse OS than those with single hits TP53 (HR= 2.5; 2-3). However, a greater difference in OS between single and double TP53 hits was observed in MDS (HR= 3.1; 2.43- 4.09) compared to AML (HR= 1.5; 1.2- 2). When we compared TP53 single hits (traditional criteria) to TP53WT in both AML (HR: 1.8 [1.4-2.5]) and MDS (HR: 1.3 [1-1.7]), TP53MTcases were associated with worse OS.
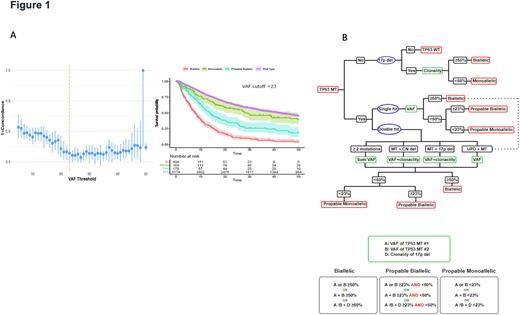
To further resolve the uncertainty of TP53MT allelic status assessment, we applied ML-driven recursive partitioning on cumulative hazard objective benchmarked to external survival data. Obligatory biallelic cases were identified among patients with: i) single TP53 hit and VAF >50%, ii) double TP53 hits with a combined VAF>50%, iii) TP53MT VAF+del17p clonality >50%. Cases with VAF <50% can be differentiated into those likely with truly monoallelic TP53MTand/or likely containing subclonal cryptic biallelic TP53 hits. We then cross-validated VAF cutoff values (Fig1-A) and applied random forest regression2 with survival as a surrogate marker for TP53 allelic status. A VAF cutoff of 23% was found to be optimal in separating monoallelic and biallelic TP53MT based on the frequency of VAF splitting criteria and decrease in error. Thus, we classified TP53MT into: A) "obligatory" biallelic (OBi), B) "probable biallelic" (PBi), and C) "probable monoallelic" (PMo). Based on this approach, 579 patients (57%) had OBi, 239 (24%) had PBi, and 192 (19%) had PMoTP53MT(Fig1-B). Of note, the OS of patients with PMo TP53MT (median OS: 29 [10-77]) was similar to that of the TP53WT (median: 42 [15-103]), p=.070). However, patients with PBi TP53MT (median OS: 14 [7-37]) had worse outcomes compared to TP53WT (p<.001). Furthermore, OS was similar when we compared AML to MDS within the OBi TP53MT. These results were recapitulated in our single cell DNA sequencing studies. In 4 patients (3 likely monoallelic , 1 likely biallelic TP53MT) a combined single cell DNA and CN analysis was performed. We found that some cells contained monoallelic TP53MT while others had indeed biallelic lesions in all cases studied. Finally, our new algorithm was able to further discriminate OS differences beyond the traditionally classified single and double TP53 hit groups. Validity of our algorithm was confirmed in an external confirmatory cohort of 92 patients.
In sum, our novel approach more accurately resolves TP53 genomic configuration and uncovers genetic mosaicism to improve the prognostic evaluation.
1
2
Disclosures
Madanat:Sierra Oncology, Stemline Therapeutics and Novartis: Membership on an entity's Board of Directors or advisory committees; BluePrint Medicines, GERON, OncLiv: Consultancy, Honoraria. Balasubramanian:Kura Oncology: Research Funding. Meggendorfer:MLL Munich Leukemia Laboratory: Current Employment. Carraway:Stemline: Speakers Bureau; Novartis: Honoraria, Speakers Bureau; Gilead: Consultancy, Membership on an entity's Board of Directors or advisory committees; AbbVie: Other: DSMB; Jazz: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Takeda: Other: DSMB; Syndax: Other: DSMB; CTI Biopharma: Consultancy, Membership on an entity's Board of Directors or advisory committees; BMS: Consultancy, Honoraria, Speakers Bureau. Mukherjee:Jazz Pharmaceuticals: Other: Principal investigator for Investigator Initiated Trials (the Institution gets the funding), Research Funding; BioPharm: Consultancy; Celgene/Acceleron: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Other: Advisor or review panel participant, Research Funding; Partnership for Health Analytic Research, LLC: Honoraria; BMS: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Other: Advisor or review panel participant, Research Funding; Aplastic Anemia and MDS International Foundation: Honoraria; Eusa Pharma: Consultancy, Other: Advisor or review panel participant; Teaching and Speaking; McGraw Hill Hematology Oncology Board Review: Honoraria, Other: Advisor or review panel participant; AbbVie: Membership on an entity's Board of Directors or advisory committees, Other: Advisor or review panel participant; Genentech: Membership on an entity's Board of Directors or advisory committees; Novartis: Consultancy, Membership on an entity's Board of Directors or advisory committees, Other: Advisor or review panel participant, Research Funding; Blueprint Medicines: Membership on an entity's Board of Directors or advisory committees, Other: Advisor or review panel participant. Sekeres:Novartis: Membership on an entity's Board of Directors or advisory committees; Takeda/Millenium: Membership on an entity's Board of Directors or advisory committees; Bristol Myers-Squibb: Membership on an entity's Board of Directors or advisory committees; Kurome: Membership on an entity's Board of Directors or advisory committees. Haferlach:Munich Leukemia Laboratory: Current Employment, Other: Part ownership. Maciejewski:Alexion: Consultancy; Apellis Pharmaceuticals: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal